Can Deep Transfer Learning Help Detect COVID-19?
Disclaimer:The information provided and referenced here is meant for Deep Learning educational purposes only. In no way is it implied that this technique should be used for the identification of COVID-19. The number of available COVID-19 chest X-rays images is too small to build a reliable model. We are using datasets from disparate sources, collected at different times with different procedures. I have no way of knowing if the image is really of a COVID-19 Chest X-ray, or some other ailment that resembles COVID-19. Consider this post an interesting use case of applying Deep Transfer Learning to a set of images for classification.
Can we use Deep Learning to analyze a set of chest X-rays of known COVID-19 patients to detect when a patient has COVID-19 and when the chest X-ray is normal?
In this blog post we will look at how we can apply Deep Learning, and in particular transfer learning via fine tuning the network. This will help discover what COVID-19 chest X-rays look like and differentiate them from normal chest X-rays or pneumonia chest X-rays.
Deep Learning and transfer learning are very involved topics and this post is more about the application of the techniques and not a tutorial on the how to develop Deep Learning models.
The goal is to demonstrate how Machine Learning techniques can be applied to a very real, and a very serious problem the world is facing. Every field will be touched by Machine Learning, and the medical field is one that will be heavily augmented by this technology. It is the hope that technology like Deep Learning will assist in the triage of patients that can be determined healthy quickly and efficiently, so doctors and clinicians can focus on those that need medical attention. Deep Learning will assist in triage to determine patients are healthy quickly and efficiently, so doctors can focus on those that need medical attention. Share on X
References
This post was inspired by a blog post by Adrian Rosebrock on his website PyImageSearch. In that blog he covers the programming details around how to apply TensorFlow and Keras to the COVID-19 image classification.
I also have a Github repo that contains the implementation for the information in this blog post. In my Github repo I build upon Adrian’s post by testing an additional deep learning model, and extending the model to predict one of 3 possible classifications; COVID, NORMAL, PNEUMONIA.
For a thorough treatment of Deep Learning for Computer Vision, I would highly recommend Adrian’s Deep Learning for Computer Vision with Python book and the PyImageSearch blogs in general.
For a general treatment of Deep Learning with Keras, I recommend the great book by Francois Chollet the developer of Keras, Deep Learning with Python. The first edition was released in 2017 and still relevant, but uses the 1.x version of TensorFlow. There is a new version being written now that will be available soon.
DataSets
Two datasets will be used for this analysis. One one source collection of chest X-Rays of COVID-19 patients hosted on Github. There other is from the Kaggle site which contains chest X-Rays of normal lung and those with pneumonia.
COVID-19 Chest X-Ray Dataset
Joesph Paul Cohen has created a GitHub repo to collect chest X-Rays of anonymized COVID-19 patients. He is also collecting other respiratory X-rays such as MERs and SARs.
From the Github repo it state:
We are building a database of COVID-19 cases with chest X-Ray or CT images. We are looking for COVID-19 cases as well as MERS, SARS and ARDS.
We will use this dataset to have a model learn what a COVID-19 chest X-Ray looks like. This dataset is updated frequently. New images were added as I was working with the dataset so check back often.
At the time of this writing, 102 COVID-19 images are available. When the blog post mentioned above was written March 16, only 25 images were available at that time.
Kaggle Chest X-Ray Images (Pneumonia)
The second dataset come from Kaggle. Kaggle is an online community of people interested in data science. It allows users to find, publish, explore, and build machine learning models around dataset made available to the public.
The dataset we will use is the Chest X-Ray Images (Pneumonia) dataset. This dataset has chest X-ray images of normal lungs as well as chest X-ray images of lungs with pneumonia.
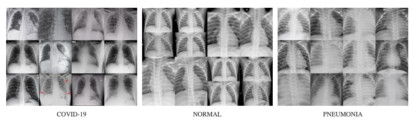
We can use this dataset to train the model on what normal chest X-rays look like. Below is a sample of the chest X-rays from each of the datasets.
Convolutional Neural Networks
Convolutional Neural Networks (CNNs) are commonly used when working with images and Deep Learning.
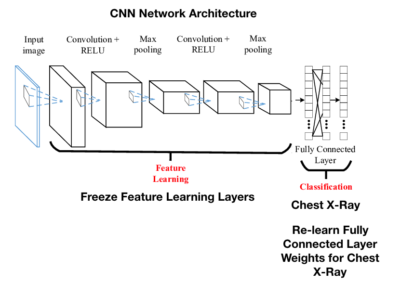
Below is a representation of the entire CNN model with a Feature learning part (CNN layers) and the classification part (FCN layers).
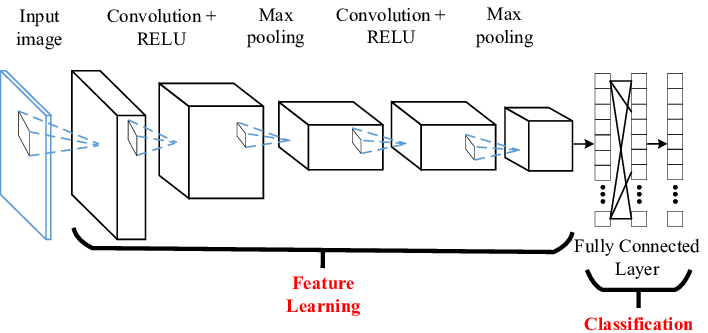
Think of a convolution as a small, 3×3 or 5×5, etc, mathematical matrix, called a kernel, that is applied to an image to alter the image in some way. We use convolutions all the time when using image processing software to sharpen or blur an image. The goal of training a CNN is to determine the values in the kernels that will alter the image in such a way as to expose features of the image that the FCN layer can use for classification.
A CNN model will be made up of some number of convolutional layers. Each layer will have a different number of kernels (small matrix we talked about), and a final fully connected network (FCN) layer that will be used to perform the actual classification step.
The initial convolutional layers, also called the convolutional base, act as a feature extraction layer. This layer is attempting to identify the features in a dataset, and in particular, for an image that might have identifying parts of the images. According to Francois Chollet the creator of Keras from his book Deep Learning with Python.
… the representations learned by the convolutional base are likely to be more generic [than the fully connected layer] and therefore more reusable; the feature maps of the convnet are presence maps of generic concepts over a picture, which is likely to be useful regardless of the conputer-vision problem at hand.
This means that the convolution layers can be trained to identify interesting features based on how the model was trained. This does imply that the model was trained on images with some commonality to the new problem.
So if your new dataset differs a lot from the dataset on which the original model was trained, you may be better off using the first few layers of the model to do feature extraction, rather than using the entire convolutional base. (Francois Chollet – Deep Learning with Python)
What is Deep Transfer Learning – Fine Tuning
There are two types of transfer learning; feature extraction and fine tuning.
Fine tuning is when the FCN layer of a CNN is removed and retrained with a new FCN layer.
The representation learned by the new fully connected network layer will be specific to the new dataset that the model is trained on, as it will only contain information about the set of possible outcomes. In our example, we are using a new fully connected neural network layer to learn how to classify COVID, NORMAL and PNEUMONIA check X-rays.
This means we can use a trained model leveraging all of the time and data it took for training the convolutional part, and just remove the FCN layer.
A properly trained CNN requires a lot of data and CPU/GPU time. If our dataset is small, we cannot start from a fresh CNN model and try to train it and hope for good results.
Applying Deep Transfer Learning
This section is meant to just scratch the surface of Deep Transfer Learning and how that is accomplished with TensorFlow and Keras. It is one thing to say, “just remove the FCN layer from the pre-configured CNN” and another to see what that means in the software.
The resources mentioned above are very good for deep treatment of transfer learning.
For this post we will look to see how to use VGG16 for transfer learning. My Github repo will use VGG16 and VGG19, and shows you how to use all both models for transfer learning. There are number of CNN architectures in the Keras library to choose from. Different CNN architectures have very different performance characteristics. ResNet architecture was very popular in 2015 timeframe. It was not included in this blog, because on the COVID-19 dataset, it did not perform well, and is not used in the model results section. The VGG architectures did perform very well.
To setup the Deep Learning models for transfer learning, we have to remove the fully connected layer. Create the VGG16 class and specify the weights as ‘imagenet’ weights and include_top=False. This will pre-initialize all of the weights to be those trained on the ImageNet dataset, and remove the top FCN layer.

The first parameter, weights is set to imagenet. This means we want to use the kernel values for all of the convolutional matrices used to train the very large ImageNet dataset. Doing so means we can leverage all of the training done previously on a huge dataset so we do not have to do that.
The second parameter, include_top is set as False. This remove the FCN layer from the VGG16 convolutional neural network.
The third parameter, input_tensor is set to the Input shape of the images.
Now that we have removed the FCN layer, we need to add our own FCN layer that wont have any weights. The new weights for the FCN layer will need to be learned by training the model on the new Chest X-ray data.

Here we start with the baseModel, which is the VGG16 model architecture, initialized with the ‘imagenet’ weights and we are going to add a new FCN layer of 64 nodes, followed by a drop out layer randomly removing 1/2 the nodes to reduce overfitting. Then, feed that into a 3 nodes output layer.
There is nothing special about this particular FCN layer architecture. Part of the task of machine learning is to experiment with different architectures to determine which is best for the problem at hand.
The 2 nodes represent the probability of a COVID-19 X-ray or a Normal X-ray.
The last step, if you recall from the picture above, is that we have to freeze the convolutional layers because we do not want to retrain that layer – we want to use the values from imagenet. When weights are adjusted, we only want the new FCN layer weights to be adjusted.

At this point the new VGG16 model is ready to train against the COVID-19 and Normal Chest X-ray data.
The VGG16 model uses the convolutional network architecture as outlined in the research paper, with all of the convolutional kernel weights already initialized with the imagenet weights. This will save a lot of time and CPU cycles.
Training the Models
My Github repo has the details of how to train the models and download the datasets so I won’t repeat that here. We will go right to the results.
Model Results
VGG16
--------- Model: vgg16 -------------
Classification Report:
precision recall f1-score support
covid 0.88 1.00 0.93 21
normal 0.78 0.86 0.82 21
pneumonia 0.93 0.70 0.80 20
accuracy 0.85 62
macro avg 0.86 0.85 0.85 62
weighted avg 0.86 0.85 0.85 62
Confusion Matrix
Predictions
covid normal pneumonia
covid 21 0 0
Actual normal 2 18 1
pneumonia 1 5 14
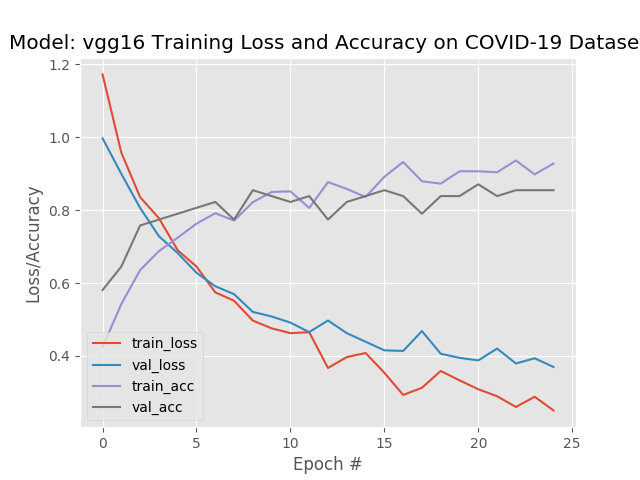
The recall, or true positive rate, is the proportion of actual positives the model classified correctly. As you can see, the recall for covid was 1.00, meaning that all of the actual COIVID-19 samples were classified correctly. However the covid precision, which is a measure of the predicted positives, is 88%. This means that 88% of the time when it predicted covid it was correct. That means 12% of the time the model predicted covid, but the actual target was normal or pneumonia.
The “confusion matrix” shows how the predictions are distributed across the possible set of categories.
A perfect recall means that the model, albeit with the limited dataset, would not generally predict someone does not have COVID-19, when they actually do.
However we can see that we have 3 samples where the model predicted the patient had COVID-19, but did not.
This is the tradeoff that has to be balanced. Would we rather have the model error with a “False Negative” (falsely state a patient does not have COVID but actually does), and release an infected person into the general population? Or error with a “False Positive” (falsely state a patient does have COVID when they do not) where the model predicts the person does have COVID-19, quarantine them, and it turns out they do not have COVID-19?
One outcome can be catastrophic and the other is a serious inconvenience.
Let’s now look at the performance of another model in the same family, VGG19
VGG19
--------- Model: vgg19 -------------
Classification Report:
precision recall f1-score support
covid 0.95 0.90 0.93 21
normal 0.86 0.90 0.88 21
pneumonia 0.85 0.85 0.85 20
accuracy 0.89 62
macro avg 0.89 0.89 0.89 62
weighted avg 0.89 0.89 0.89 62
Confusion Matrix
Predictions
covid normal pneumonia
covid 19 0 2
Actual normal 1 19 1
pneumonia 0 3 17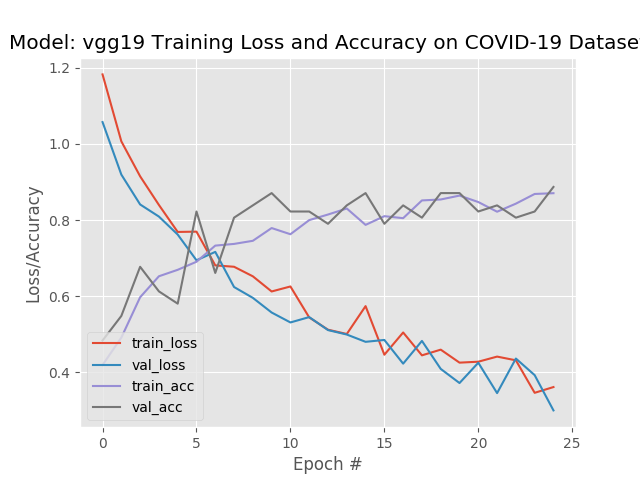
VGG19 had a slightly better average accuracy, but a lower recall. In this situation I would tend to go with the model that produced the higher recall.
Test on Kaggle Test Images
Let’s test the models on normal and pneumonia chest X-ray images from the Kaggle dataset that the models have never seen. Ideally we would also have COVID-19 images that we held out but we used all of the limited COVID-19 images to train the model and you should test on images that were used for training.
We are looking for a measure of how well the model does in predicting NON-COVID19, on “healthy” X-rays from the Kaggle dataset.
My Github repo has a script to run the predictions. We will just review the results here.
Kaggle NORMAL Chest X-Rays
For this test we are taking all NORMAL test chest X-rays and measuring how the model performed.
Model: ./models/best-vgg16-0402-model.h5 Dataset: /Volumes/MacBackup/kaggle-chest-x-ray-images/chest_xray/test/NORMAL normal Accuracy: 0.68 COVID-19 %: 0.15 Normal %: 0.68 Pneumonia %: 0.16 NON-COVID-19 %: 0.84
The best VGG16 model was able to predict NORMAL chest X-rays, 68% of the time. But if we are interested in more of a COVID, NOT-COVID, and if we combine the Normal and Pneumonia predictions, we are up to 84% of the time, rounding errors not withstanding.
Kaggle PNEUMONIA Chest X-Rays
Model: ./models/best-vgg16-0402-model.h5 Dataset: /Volumes/MacBackup/kaggle-chest-x-ray-images/chest_xray/test/PNEUMONIA pneumonia Accuracy: 0.85 COVID-19 %: 0.04 Normal %: 0.12 Pneumonia %: 0.85 NON-COVID-19 %: 0.97
The best VGG16 model was able to predict PNEUMONIA 85% of the time, and when combined with Normal the NON-COVID-19 prediction is close to 97%. Again, rounding errors not withstanding.
Recap
The purpose of this post was to demonstrate how we can use Deep Transfer Learning for image classification. The images we used were those of COVID-19 chest X-Rays.
Transfer learning allows us to leverage the weights of pre-trained models so we do not have to retrain every model from scratch. For situations where we do not have a lot of training data, using transfer learning is an absolute necessity.
The idea behind transfer learning is to use the convolutional layers of an existing model architecture, with weights from models training on huge datasets. These convolutional layers become the feature extractors for new images that the model was never trained on. Using the extracted features, we use a new fully connected network layer to learn the classification for our problem. In this case whether a chest X-Ray showed COVID-19, was Normal or Pneumonia. Training the fully connected network layer requires much less time and data, although the more data the better.
Lastly this post looked at how model performance as measured by the a test holdout set of Normal and Pneumonia chest X-Rays.
You can find the source code used for this post on my Github repo.
Final Disclaimer
As mentioned earlier, this post is covering a topic that is impacting all of us to some degree. This post is meant to use available datasets to show how to perform image classification using Deep Transfer Learning. It is not meant to imply using these techniques for any medical purpose.